Part III:
Arlin: Here are the alignments that I have been promising to do for you. Here is what I did.
1A. For you, the quickest way to do this (if you want to check my work, or to look at gene sequence yourself) is to access the SWISS-PROT database from your Macintosh using Turbogopher. Here are the Mac Turbogopher attributes for our bookmark link to the SWISS-PROT database:
+INFO: 7Search SWISS-PROT (Release 31) 7/gopherlib/indices/swiss/index gopher.nih.gov 70
1B. Now query the SWISS-PROT database with "<organism>" and "<globin>", where:
<organism> = { chicken, cow, horse, minke whale, mouse, rat, tuna }
<globin> = { hemoglobin, myoglobin }
For each query, there will be one or more hits. Pick one hit for each myoglobin query. For each "hemoglobin" query, pick an "alpha" hemoglobin and a "beta" hemoglobin.
Then I aligned the sequences.
2A. Cut and paste the amino acid sequences into a flat text file in the format of the attached data below (i.e., a right cursor followed by a brief name, followed by the sequence on a new line).
2B. Run the sequences through a multiple alignment program. I used two different programs, one called "multalin" and the other called "clustalv". Both of these programs are freely available to scientists.
The alignment is below. I'll describe how this relates to hypothesis-testing in the following message. For now, just note that the names given at the beginning of each line use the following convention:
AHB=alpha-hemoblogin
BHB=beta-hemoglobin
MB=myoglobin
CHIX=chicken
COW=cow
MOUS=mouse
HORS=horse
MINK=minke whale (the program chops the name)
RAT=rat
TUNA=tuna
The amino acid code is A=alanine, C=cysteine, D=aspartate, E=glutamate, and so on.
So the first line means that the sequence of alpha-hemoglobin of chicken is VLSAADK..., that is, Valine-Leucine-Serine-Alanine-Alanine-Aspartate-Lysine...
[ 1 10 20 30 40 50 60 ] AHBCHIX VLSAADKNNVKGIFTKIAGHAEEYGAETLERMFTTYPPTKTYFPHF-DLSH-----GSA AHBCOW VLSAADKGNVKAAWGKVGGHAAEYGAEALERMFLSFPTTKTYFPHF-DLSH-----GSA AHBMOUS VLSGEDKSNIKAAWGKIGGHGAEYGAEALERMFASFPTTKTYFPHF-DVSH-----GSA AHBHORS VLSAADKTNVKAAWSKVGGHAGEYGAEALERMFLGFPTTKTYFPHF-DLSH-----GSA AHBMINK VLSPTDKSNVKATWAKIGNHGAEYGAEALERMFMNFPSTKTYFPHF-DLGH-----DSA AHBRAT VLSADDKTNIKNCWGKIGGHGGEYGEEALQRMFAAFPTTKTYFSHI-DVSP-----GSA AHBTUNA TTLSDKDKSTVKALWGKISKSADAIGADALGRMLAVYPQTKTYFSHWPDMSP-----GSG BHBCHIX VHWTAEEKQLITGLWGKV--NVAECGAEALARLLIVYPWTQRFFASFGNLSSPTAILGNP BHBCOW MLTAEEKAAVTAFWGKV--KVDEVGGEALGRLLVVYPWTQRFFESFGDLSTADAVMNNP BHBMINK VHLTAEEKSAVTALWAKV--NVEEVGGEALGRLLVVYPWTQRFFEAFGDLSTADAVMKNP BHBHORS VQLSGEEKAAVLALWDKV--NEEEVGGEALGRLLVVYPWTQRFFDSFGDLSNPGAVMGNP BHBMOUS VHLTDAEKAAVSCLWGKV--NSDEVGGEALGRLLVVYPWTQRYFDSFGDLSSASAIMGNA BHBRAT VHLTDAEKAAVNGLWGKV--NPDDVGGEALGRLLVVYPWTQRYFDSFGDLSSASAIMGNP BHBTUNA VEWTQQERSIIAGFIANL--NYEDIGPKALARCLIVYPWTQRYFGAYGDLSTPDAIKGNA MBCHIX GLSDQEWQQVLTIWGKVEADIAGHGHEVLMRLFHDHPETLDRFDKFKGLKTPDQMKGSE MBCOW GLSDGEWQLVLNAWGKVEADVAGHGQEVLIRLFTGHPETLEKFDKFKHLKTEAEMKASE MBHORSE GLSDGEWQQVLNVWGKVEADIAGHGQEVLIRLFTGHPETLEKFDKFKHLKTEAEMKASE MBMINKE VLSDAEWHLVLNIWAKVEADVAGHGQDILIRLFKGHPETLEKFDKFKHLKTEAEMKASE MBMOUSE GLSDGEWQLVLNVWGKVEADLAGHGQEVLIGLFKTHPETLDKFDKFKNLKSEEDMKGSE MBMRAT GLSDGEWQLVLNVWGKVEGDLAGHGQEVLIKLFKNHPETLEKFDKFKHLKSEDEMKGSE MBTUNA ----ADFDAVLKCWGPVEADYTTMGGLVLTRLFKEHPETQKLFPKFAGI-AQADIAGNA [ 1 10 20 30 40 50 60 ] [ 70 80 90 100 110 120 ] AHBCHIX QIKGHGKKVVAALIEAANHIDDIAGTLSKLSDLHAHKLRVDPVNFKLLGQCFLVVVAIHH AHBCOW QVKGHGAKVAAALTKAVEHLDDLPGALSELSDLHAHKLRVDPVNFKLLSHSLLVTLASHL AHBMOUS QVKGHGKKVADALASAAGHLDDLPGALSALSDLHAHKLRVDPVNFKLLSHCLLVTLASHH AHBHORS QVKAHGKKVGDALTLAVGHLDDLPGALSNLSDLHAHKLRVDPVNFKLLSHCLLSTLAVHL AHBMINK QVKGHGKKVADALTKAVGHMDNLLDALSDLSDLHAHKLRVDPANFKLLSHCLLVTLALHL AHBRAT QVKAHGKKVADALAKAADHVEDLPGALSTLSDLHAHKLRVDPVNFKFLSHCLLVTLACHH AHBTUNA PVKAHGKKVMGGVALAVTKIDDLTTGLGDLSELHAFKMRVDPSNFKILSHCILVVVAKMF BHBCHIX MVRAHGKKVLTSFGDAVKNLDNIKNTFSQLSELHCDKLHVDPENFRLLGDILIIVLAAHF BHBCOW KVKAHGKKVLDSFSNGMKHLDDLKGTFAALSELHCDKLHVDPENFKLLGNVLVVVLARNF BHBMINK KVKAHGKKVLASFSDGLKHLDDLKGTFATLSELHCDKLHVDPENFRLLGNVLVIVLARHF BHBHORS KVKAHGKKVLHSFGEGVHHLDNLKGTFAALSELHCDKLHVDPENFRLLGNVLVVVLARHF BHBMOUS KVKAHGKKVITAFNDGLNHLDSLKGTFASLSELHCDKLHVDPENFRLLGNMIVIVLGHHL BHBRAT KVKAHGKKVINAFNDGLKHLDNLKGTFAHLSELHCDKLHVDPENFRLLGNMIVIVLGHHL BHBTUNA KIAAHGVKVLHGLDRAVKNMDNINEAYSELSVLHSDKLHVDPDNFRILGDCLTVVIAANL MBCHIX DLKKHGATVLTQLGKILKQKGNHESELKPLAQTHATKHKIPVKYLEFISEVIIKVIAEKH MBCOW DLKKHGNTVLTALGGILKKKGHHEAEVKHLAESHANKHKIPVKYLEFISDAIIHVLHAKH MBHORSE DLKKHGTVVLTALGGILKKKGHHEAELKPLAQSHATKHKIPIKYLEFISDAIIHVLHSKH MBMINKE DLKKHGNTVLTALGGILKKKGHHEAELKPLAQSHATKHKIPIKYLEFISDAIIHVLHSRH MBMOUSE DLKKHGCTVLTALGTILKKKGQHAAEIQPLAQSHATKHKIPVKYLEFISEIIIEVLKKRH MBMRAT DLKKHGNTVLTALGGILKKKGQHAAEIQPLAQSHATKHKIPIKYLEFISEAIIQVLQSKH MBTUNA AISAHGATVLKKLGELLKAKGSHAAILKPLANSHATKHKIPINNFKLISEVLVKVMHEK- [ 70 80 90 100 110 120 ] [ 130 140 150 154 ] AHBCHIX PAALTPEVHASLDKFLCAVGTVLTAKYR AHBCOW PSDFTPAVHASLDKFLANVSTVLTSKYR AHBMOUS PADFTPAVHASLDKFLASVSTVLTSKYR AHBHORS PNDFTPAVHASLDKFLSSVSTVLTSKYR AHBMINK PAEFTPSVHASLDKFLASVSTVLTSKYR AHBRAT PGDFTPAMHASLDKFLASVSTVLTSKYR AHBTUNA PKEFTPDAHVSLDKFLASVALALAERYR BHBCHIX SKDFTPECQAAWQKLVRVVAHALARKYH BHBCOW GKEFTPVLQADFQKVVAGVANALAHRYH BHBMINK GKEFTPELQAAYQKVVAGVANALAHKYH BHBHORS GKDFTPELQASYQKVVAGVANALAHKYH BHBMOUS GKDFTPAAQAAFQKVVAGVATALAHKYH BHBRAT GKEFTPCAQAAFQKVVAGVASALAHKYH BHBTUNA GDAFTVETQCAFQKFLAVVVFALGRKYH MBCHIX AADFGADSQAAMKKALELFRNDMASKYKEFGFQG MBCOW PSDFGADAQAAMSKALELFRNDMAAQYKVLGFHG MBHORSE PGNFGADAQGAMTKALELFRNDIAAKYKELGFQG MBMINKE PAEFGADAQAAMNKALELFRKDIAAKYKELGFQG MBMOUSE SGDFGADAQGAMSKALELFRNDIAAKYKELGFQG MBMRAT PGDFGADAQGAMSKALELFRNDIAAKYKELGFQG MBTUNA -AGLDAGGQTALRNVMGIIIADLEANYKELGFSG [ 130 140 150 154 ]
Arlin: You may have noticed in the sequence alignment of globins that the alpha hemoglobin chains had similar gaps (the "-"s) and similar amino acid sequences. Likewise with the beta-hemoglobin and the myoglobin sequences. From the point of view the descent-with-modification theory, this similarity is due to common ancestry -- each of the three types of globins arose from a different common ancestor. If the beta-hemoglobin common ancestor had a certain set of differences from the alpha-hemoglobin common ancestor, then subsequent hemoglobins will inherit these differences.
From the point of view of the spontaneous-generationist or creationist or Senapatheist, the similarities are due to functional demands. Alpha hemoglobins must have particular sequences in order to fulfill their roles, while beta hemoglobins and myoglobins must have a different type of sequence. Therefore, it is clear that god would have created alpha hemoglobins with different sequences than myoglobins. From the Senapatheistic perspective, only certain sequences can function as alpha-hemoglobin sequences, therefore seed cells must have certain alpha-hemoglobin sequences and different beta-hemoglobin sequences.
There are some subtleties that I am ignoring here (e.g., god wouldn't really make rat myoglobin different from cow myoglobin; seed cells could not be under selection for hemoglobin function, given that they don't have lungs, blood, muscles, etc.), but basically we are unable to resolve these different positions on the basis of global comparisons alone.
However, the implications of these different views are very different with regard to the detailed pattern of differences. In particular, the descent-with-modification theory predicts that the arrangement of species in a phylogenetic tree inferred for alpha-hemoglobin will tend to look like the tree inferred from beta-hemoglobin sequences, and like the tree inferred from myoglobin sequences. The reason for this is that (if organisms only rarely share their genes with each other) each family of genes will have evolved through the same branching pathway of ancestor-descendant relationships. In Senapatheism, species are born INDEPENDENTLY, and their genomes are assembled from random sequences, therefore the alpha-hb, beta-hb and myoglobin trees will have nothing particular in common. In creationism, there are no adequate grounds for prediction, unless one believes in biblical creationism, in which case the bible offers clues as to which groups of organisms were created on the same day -- presumably, god would have used the same templates as much as possible for organisms created on the same day.
So, here is a clear chance for creationism or for Senapathy's theory to strike a crushing blow to the Darwinian hegemony. If the trees for myoglobin and for alpha- and beta-hemoglobin are independent (i.e., as in Independent Birth of Organisms), then Senapathy will be vindicated and Darwin will be left with egg on his face. Let me remind you that Senapathy fully believes that this is what will happen. He said as much on sci.bio.evolution in his response to the posting in which I first described this "congruence" test.
We can make the trees whenever you are ready to make a prediction. There is publicly available software at:
http://genetics.washington.edu/phylip.html
For the Macintosh, you can download this 1-megabyte file:
ftp://genetics.washington.edu/pub/phylip/phylip.sea.hqx
and I can show you how to use it (you will need a C compiler).
Jeff: What is a "gap" in the chain?
Arlin: Not all globin chains are the same length. When two sufficiently different globin sequences are aligned relative to each other, so that similar residues are matched, there are also places where one chain has to "stretched" or "compressed" relative to each other to make the alignment work. This is presumably due to the accumulation of deletions and insertions in the gene sequences.
For instance consider the problem of aligning the first 55 or so residues of the alpha and beta hemoglobins of chicken. They have about 18% identical positions when aligned (much greater than 5%, the random chance of a match given that there are 20 different amino acids). However, to get this match, we have to introduce 4 gaps, 3 in the alpha sequence and 1 in the beta sequence:
AHBCHIX V-LSAADKNNVKGIFTKIAGHAEEYGAETLERMFTTYPPTKTYFPHF-DLSH-----GSA
BHBCHIX VHWTAEEKQLITGLWGKV--NVAECGAEALARLLIVYPWTQRFFASFGNLSSPTAILGNP
* * * * * * *** * ** * * * ** *
There is no physical "gap" in the protein chain. If you collect some mouse blood and isolate the hemoglobin chains, each alpha or beta chain will have 14X residues, with each residue linked to the next residue by a peptide bond. The "gap" is a conceptual one, created in order to improve the alignment score for a bunch of sequences in a computer. However, this conceptual gap does have a physical implication, suggesting (as it does) that gene sequences undergo deletions and insertions over time.
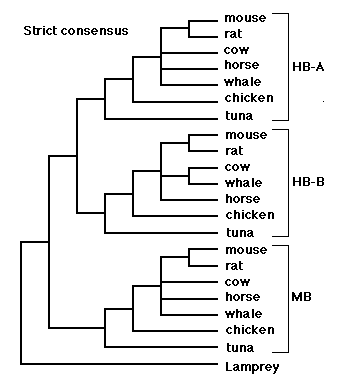
Arlin: Here is a phylogenetic tree derived from the above sequences. I added an "outgroup" sequence from the lamprey. This allows a root to be placed on the tree. I searched for the most parsimonious tree using the Macintosh program "PAUP" (Phylogenetic Analysis Using Parsimony). Twenty-four equally parsimonious trees were found before I stopped the search (out of boredom). The figure shows a "strict consensus" tree, that is, a tree with only those branches that were agreed upon by all 24 equally parsimonious trees recovered by the PAUP search. The trees differed only slightly, regarding the relative placement of rodents, horse, cow and whale.
Because the trees differ from each other, a "strict consensus" tree will contain one or more "polytomies" or "polychotomies" (as opposed to "dichotomies"), that is, nodes or forks in the tree that have more than two prongs coming out of them. The polytomy expresses the fact that the data cannot easily be resolved into a single dichotomously branching tree (with more data, it might have been possible to resolve these nodes). It's like a foot race, where we can assume a priori that contestant A must have finished either before or after contestant B, yet sometimes the judges at the race are incapable of distinguishing the order, and a tie must be declared. A polytomy in a tree is a bit like a tie. We know that the rooted order of taxa A, B and C is either
(A (B,C)), (C (A,B)) or (B (A,C)),yet we do not know which one is correct. That ambiguity can be represented with the polychotomous tree (A, B, C).
Anyway, the consensus tree splits the sequences into three sequence families corresponding to alpha-hemoglobin chains, beta-hemoglobin chains, and myoglobin chains. The important question with regard to falsifying independent birth is the question of congruence between the different sequence families. In fact, the within-sequence-family relationships are entirely congruent within the limits of resolution of the method. Within each sequence family, the tuna (fish) is an outgroup to the chicken+mammals; within the chicken+mammal group, the chicken is an outgroup to the mammals. The within-group relationships of mammals are not completely resolved, except that the rat and mouse are always united (they are both members of the mammalian order "rodentia").
The significance of congruence was explained earlier in our correspondence, and also in sci.bio.evolution, in a posting that is reproduced on your web page under the heading "From Arlin Stoltzfus in bionet.molbio.evolution". Basically, if taxa A, B, and C have genomes that were independently and randomly assembled, then a tree of gene X will not look like a tree of gene Y from A, B and C, since each is just a collection of sequences that vary randomly. Congruence, a tendency for the trees for gene X and gene Y to match each other, is incompatible with the independent birth theory. As Senapathy says:
Even if [the Independent Birth theory is true], still we can "construct" a "phylogenetic tree" for the molecules such as the 16S rRNA. Such trees are only false trees, and do not represent the reality. Please note that each gene included in the different genomes could change into its many normal variants with sequence changes. If we analyze these RNA sequences with the assumption of independent genome assembly, I am sure we shall see that the results will fit the new theory.
That is, if the independent birth theory were true, the trees for each different type of chain would be random noise, representing variation around a mean ("normal", he says). It would be possible to make a tree (as I explained in sci.bio.evolution, and as Senapathy agrees in the passage quoted above), but the trees do not have meaning and will not show congruence. That is, if the independent birth theory is true, the alpha-hemoglobin tree will not give us any clue as to what the beta-hemoglobin tree looks like, since both the alpha and beta families will just be collections of random sequences, independently assembled:
They [i.e., the sequences of genes] changed randomly in the different genomes without losing the essential structure or function, only changing to their normal variants. These normal sequence variations are those that evolutionary molecular biologists now use to "construct" a misleading phylogenetic tree.
So, you tell me: do "the results fit the new theory" as Senapathy suggests? Does the alpha-hemoglobin tree tell us NOTHING about the beta-hemoglobin tree, since each is just a meaningless tree made from random noise? You know that the answer is negative. The tree for one type of molecule can be used to predict the pattern of branching in the tree for another type of molecule -- this is NOT RANDOM, NOT INDEPENDENT.
By the way, this result can't be explained by supposing that the common branching pattern is due to restrictions in the range of "normal" (Senapathy's word, not mine) variation determined by common lifestyles selected from the primordial pond. That is, maybe rat and mouse sequences are more similar (and group together in the tree) because rat and mouse are both furry little seed-eating warm-blooded animals. Fair enough, for rat and mouse. However, please note that whale sequences tend to be more similar to horse sequences than to tuna sequences. This is true for myoglobin, alpha-hemoglobin, and beta-hemoglobin. If the similarities were due to restrictions in the range of "normal" (Senapathy's word, not mine) determined from the primordial pond, then whale sequences would tend to be more similar to tuna sequences than to horse sequences.
Jeff: You have done a lot of work, and I appreciate it very much. You ask: "do the results fit the new theory as Senapathy suggests?"
Unfortunately, I must disappoint you by saying, yes, the results do fit Senapathy's theory. Or, more to the point of your presentation, the results do not disprove his theory.
Two trees, three trees, many trees, IF THEY ARE CONGRUENT WITH EACH OTHER, DISPROVE INDEPENDENT BIRTH, whether it's creationism or Senapatheism.
Saying that any two or more trees, if congruent, automatically disproves Senapathy's theory is overlooking the reuse of genes in the random assembly of seed cells, and that some genes could easily be linked so their reuse maintains and propagates their variations. For example, if two genes are located near each other in the DNA, then it should be no surprise when the two genes are reused together, complete with their accumulated sequence variations plus, perhaps, some new variations. Even if the genes are not linked though physical proximity, you cannot disprove independent births by looking at genomes that were assembled so close together in time. Random assembly does not mean the DNA went through an egg beater. You are confusing the terms "random" and "independent."
Taken to its extreme, your argument that two similar trees disproves independent births is like saying that finding two different organisms with four legs, two eyes, and a nose also disproves independent births. You forget that Senapathy's theory actually predicts similarities, and it allows sequence differences as well as congruence.
What we should be looking at are not similar proteins for similar functions in closely-related organisms, but different proteins for similar functions in distant organisms, and then figure out how those proteins could be related. Senapathy has recently written quite a bit about this subject as a test of his theory ("test 1").
Please take on these challenges:
Jeff: (reprise) Your three trees agree about the relationship between tuna, chicken, and everything else (rat, mouse, whale, cow, and horse), but there is nothing convincing about the "everything else" groupings. You have chosen a set of organisms that are so close in terms of either evolution or independent births that the two theories should display similar relationships in many areas.
Arlin: The prediction based on Senapathy's assumption that genes were independently assembled was that relationships would be "meaningless," to quote Senapathy himself. An alternative hypothesis is that the relationships are congruent due to descent with modification. In fact, the relationships that are resolved ARE congruent, formally excluding the possibility that all genes were independently assembled from random sequences. Please admit this before you start objecting. For the ones that weren't resolved, the question of congruence obviously can't be answered. If I had a theory that the satellites of Mars were coated with chocolate, and I presented spectrographic data that suggested this, would you object that there were some satellites that were so small that I couldn't resolve them with my telescope, therefore my theory must be wrong? No. Lack of resolution in a part of the tree is irrelevant to the question, and does not relieve you of the obligation to admit that independent random assembly of sequences is formally excluded.
(Quoting Jeff) I should have called this to your attention earlier, but the globin proteins you chose to compare are only present in a very, very few organisms. Even within the vertebrates, you are looking at a minuscule set of organisms that have obvious relationships under either theory. This closeness does not allow us to discriminate between the theories.
Arlin: Obvious relationships? Again, I wonder what sort of "relationships" are allowed under a theory that claims that organisms are INDEPENDENTLY assembled from random sequences. Close or not close, the data indicate clearly that alpha-HB, beta-HB and myoglobin genes TRAVEL THROUGH EVOLUTION TOGETHER, following a process of descent with modification. They did not follow separate evolutionary paths, but were tied together in the same genome, through a process of descent with modification.
(Quoting Jeff) You have greatly misunderstood Senapathy's comments that you quoted above. He said absolutely nothing about "random noise." A "normal variant" is not a statement about a normal probability distribution -- he is referring to codon and amino acid degeneracies. He never said the alpha- and beta-hemoglobin trees will be different or unrelated.
Arlin: Yes, he does say they are unrelated. UNRELATED. INDEPENDENTLY ASSEMBLED. RANDOM SEQUENCES.
Responses to your challenges:
Under evolution, a fish's tail does not IN PRINCIPLE have to be related to a monkey's tail. A butterfly's wing does not IN PRINCIPLE have to be related to a bird's wing. Things that look similar on the surface do not necessarily have to be related deep-down. As it turns out, IN FACT, the fish tail and the monkey tail ARE related deep-down, but the bird's wing and butterfly wing are, IN FACT, NOT. It is no "challenge" to any theory of evolution that I know of to find different non-homologous structures doing somewhat similar things. Why would this be a challenge? However, homology of any kind, a deep-down relationship of any kind, is a serious challenge to a theory of spontaneous generation. It is a challenge because homology suggests "relatedness" and there is no deep-down systematic relatedness between organisms under a theory of spontaneous generation.
I don't know. I doubt that Darwin's thoughts on this subject would be valuable.
(b) How did it form?
I don't know.
However, even if I don't know the answer to a) and b), this does not cause me to abandon principle and seek absurd magical explanations that don't make sense. When some people see an ancient pyramid or giant statue, and cannot conceive how "primitive" peoples could have built it, they dream up dramatic theories in which space-travelers landed on earth and built monuments. In order to make an improbable thing seem more "likely" they break the rules and posit all sorts of heretofore-unseen preconditions. Other people insist on taking the view that known facts and laws can be brought to bear on the problem, and instead of inferring an alien race of superior beings, they infer that these "primitive" peoples must have had both ingenuity and an incredible socio-political organization that allowed hundreds of workers to toil for decades on projects that did not have an obvious benefit and did not put food directly in their mouths (the only modern equivalents would be medieval cathedrals, or something like the NASA space program).
That is to say, some people imagine cranes, and others imagine sky-hooks.
(c) If there was one or two original organisms, why not three, a hundred, or many millions of "first" organisms?
Indeed, why not a few, or a dozen, or a million?
(d) Why is the thought of a few million original organisms so hard to accept?
A few million organisms arising by natural means, including templating, hypercycles, natural selection is not hard to accept, in principle; a few million organisms arising by spontaneous assembly from random DNA sequence is rubbish, magical, sky-hook explanation.
BTW, it is clear that there were not a million lineages that left descendants today -- otherwise why would there be a single genetic code for all cellular organisms (with the only exceptions being derived conditions)? There are billions of species today, but in 4 billion years, only a few of them will have living descendants. Many ancient organisms, but only a few ancestors. There is a difference.
Arlin: I asked you to give me a prediction beforehand. I waited for two months, but you didn't do it. I said that presenting the trees to you before coming to an agreement about predictions would be like taking O.J. to trial knowing that the jury thought that victims blood on the defendants hands, clothes and vehicle was ambiguous evidence open to a variety of interpretations. Now I find that you believe congruence to be ambiguous evidence open to a variety of interpretations, and I cannot agree with this. Congruence constrains one to propose a relationship between traits in different organisms, to the effect that those traits evolve in concert along a branching pathway of divergence. There is no alternative.
Jeff: I cannot speak for Senapathy, and I won't set limits on a test of his theory -- that is for him to do. I told you that long ago. I wanted to see the details and methodology behind the congruence tests, and that is what you very carefully showed me. And there is an "alternative," but you refuse to understand it. It is obvious to me now that you have not studied Senapathy's theory. There is no reason why congruence could not come from the pond.
Arlin: (reprise) The prediction based on Senapathy's assumption that genes were independently assembled was that relationships would be "meaningless," to quote Senapathy himself. ...the relationships that are resolved ARE congruent, formally excluding the possibility that all genes were independently assembled from random sequences. Please admit this before you start objecting.
Jeff: "Meaningless" in terms of generating an evolutionary tree. The tree you see is a "false" tree (to quote Senapathy himself). You are taking the term "independent" to mean that there can be absolutely no relationship between seed cells and organisms from the pond -- and that is not how Senapathy is using that word. You have not read and understood the theory.
Arlin: (reprise) Yes, he does say they are unrelated. UNRELATED. INDEPENDENTLY ASSEMBLED. RANDOM SEQUENCES.
Jeff: If you mean "unrelated" in the living sense, yes. But there is a relatedness in the non-living pond sense, just as the computer on my desk is related to the computer on your desk. They share common designs -- the CRT, the CPU, the memory and I/O, are all based on previous designs (reused) and I could trace the ancestor of your computer back to the ancestor of mine (both being Macs), and there is a link to the PC as well. Our computers have reused and unique features. You could construct a "tree" showing our computers' descent based on the similarities and congruence of a set of similarities. If you did not know that machines could not reproduce, you would no doubt say they descended from one another. But, I could show you that the machines were designed "independently" and have some "random" characteristics (new features not present in other machines). Uninterested observers would agree with elements of both theories.
Arlin: Ah, now we may be coming to an agreement. As I mentioned, the data on congruence of genes DOES constrain one to propose that genes evolve in concert along a branching pathway, through hereditary descent with modification. If you wish to believe that all of this descent with modification takes place in a pond, as opposed to within cells, there is nothing in the molecular data that we have discussed (yet) that would prevent you from believing this.
Jeff: I have never argued that congruence data would falsify (or prove) descent from a common ancestor. On the other hand, your entire presentation here about sequences and congruence is based on your mistaken belief that congruence falsifies the independent birth theory. That is not that case. You have misunderstood the usage of the word "independent" in the new theory because you have been basing your understanding of the theory on the wrong book -- you have been reading the dictionary, not Senapathy's book.
Arlin: (reprise) BTW, it is clear that there were not a million lineages that left descendants today -- otherwise why would there be a single genetic code for all cellular organisms (with the only exceptions being derived conditions)?
Jeff: Why should there be multiple codes? There is a common chemistry in the pond. There are zillions of things we can conceive of and ask "why not?" That will not tell us the truth about what is.
Jeff: Can you tell me the locations of the three globin genes that you used in the sequences (alpha- & beta-hemoglobin and myoglobin in chicken, cow, mouse, horse, whale, rat, and tuna)? How close are they to each other in the DNA, and how much research has been done in comparing the locations of homologous genes? I am interested in this from the point of view of reuse and congruence.
Arlin: OK, one last try to explain congruence and why it is not explainable by the "resampling" theory of Mattox.
In general, there are three facts about congruence that are inconsistent with the explanation you have proposed.
.----- species A, gene X
.--------|
| `----- B, gene X
|
|-------------- C, gene X
|
|-------------- D, gene X
|
|-------------- E, gene X
|
|-------------- F, gene X
|
`-------------- G, gene X
That is, A and B are close for the gene X sequence because it was resampled from the pond, whereas there is no obvious relationship for the other sequences of gene X. According to the theory that genomes are assembled independently and randomly, but with occasional re-sampling from the pond, you might see this for another locus, gene Y:
.-------------- species A, gene Y
|
|-------------- B, gene Y
|
|-------------- C, gene Y
|
| .----- D, gene Y
|--------|
| `----- E, gene Y
|
|-------------- F, gene Y
|
`-------------- G, gene Y
That is, in this case the same gene was resampled twice, but the recipient genomes were D and E rather than A and B. Or the trees might be the same if gene X and gene Y are tightly linked, as you have suggested. But none of this is the same as the phenomenon of congruence that extends to nested hierarchical relationships, i.e.,
( tuna ( chicken ( horse, cow, whale (rat, mouse) ) ) )NOT just
( tuna, chicken, horse, cow, whale (rat, mouse) )That is, only rat and mouse being close, and everything else showing the same degree of relatedness.
This is not science. Science is being able to say what is consistent with the theory BEFORE SEEING THE DATA. Can you do this? I'm giving you one more chance. Will the hierarchical pattern of phylogeny for a set of taxa
{ A, B, C }and a set of genes
{ X, Y, Z }tend to match even if the taxa are as deeply separated as different animal phyla, or even different kingdoms (i.e., not 'just' vertebrates)? If the genes are unlinked and not similar in function?
[ar31]
Jeff: What you see as an apparent change of attitude by me (after seeing the data) is simply the manifestation of my prior stated refusal to specify a test of Senapathy's theory on his behalf. I don't know enough about the underlying molecular biology to specify a fair test, you had prior knowledge of the data before we started (otherwise, why would you have done it?), and, more importantly, it's not my theory to prove, it's his.
You want a test of the independent birth theory to rest on congruence, but I still feel that congruence could be produced by the mechanisms in the pond (as well as common descent). You vigorously disagree.
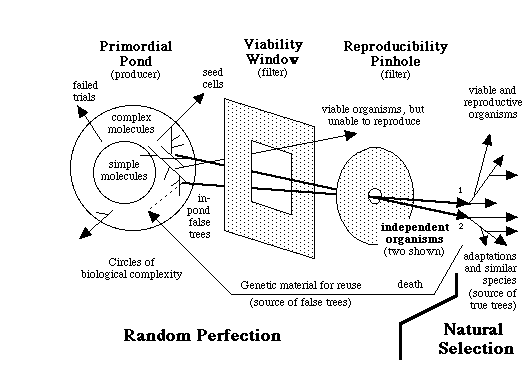
 On four occasions (three on this page alone and once last spring), you referred to figure 8.3, saying "there is no comparable concept of systematic 'closeness' in the diagram on page 311, because the theory underlying it permits of no systematic closeness of two species" and that it "shows in clear black-and-white (both in the diagram and in the caption) that different vertebrates received different genes from the pond." Here again, you misunderstand the theory, and you are misconstruing that figure -- it does not show that there are no common genes or relatedness in the output of the pond. Figure 8.3, like the fancy color cover photo, is just a simplified depiction of creatures coming from the pond. The caption says "different organisms could be derived," not that each organism has a completely different set of genes.
On four occasions (three on this page alone and once last spring), you referred to figure 8.3, saying "there is no comparable concept of systematic 'closeness' in the diagram on page 311, because the theory underlying it permits of no systematic closeness of two species" and that it "shows in clear black-and-white (both in the diagram and in the caption) that different vertebrates received different genes from the pond." Here again, you misunderstand the theory, and you are misconstruing that figure -- it does not show that there are no common genes or relatedness in the output of the pond. Figure 8.3, like the fancy color cover photo, is just a simplified depiction of creatures coming from the pond. The caption says "different organisms could be derived," not that each organism has a completely different set of genes.
Let's take a trip. Here is the full caption to figure 8.3:
"The use of the same genetic code and genetic machineries in multitudes of different organisms independently born in the primordial pond." The genetic code and the genetic machineries such as transcription, splicing, and translation systems had been already established in the primordial pond before any living cells were formed. This pool also contained the DNA-recombination enzymes, such as DNA ligases found in today's living cells, that could recombine different pieces of DNA to help form many genomes. Thus, the primordial pond was a common pool of code, genetic machineries, and genes, from which different organisms could be derived. Consequently, they all used the same code and genetic machineries, but different sets of genes and, more importantly, different developmental genetic pathways leading to distinct independently born organisms.Now, look at figure 84, on page 314. This more clearly shows the relatedness or closeness of organisms. The organisms share common genes, and they have unique genes, too. To wit:
"Independent birth of multitudes of organisms by random assembly of genes from the Universal Gene Pool (UGP) in the primordial pond." The UGP contains myriads of genes for various biochemical functions and multiple copies for each gene. Random assembly of these genes led to multitudes of independent genomes leading to mostly meaningless multicellular masses, and the rare meaningful organisms. Yet these processes resulted in numerous distinct viable creatures. Each successful genome had a unique set of genes and a unique developmental genetic pathway. This resulted in the different genomes having a subset of common genes, a subset of similar but distinct genes, and a subset of unique genes. The genomes of various organisms were assembled separately into different zygote-like "seed cells," giving rise to the independent birth of many organisms directly from the primordial pond. Parts of successful genomes were included in newer genomes being assembled from the UGP, thereby making it easier to assemble newer genomes resulting in some organismal similarity (see also figure 8.8).Again, note that saying "each successful genome had a unique set of genes" does not mean that each genome was completely different, lacking any relatedness.
And, on to figure 8.8 on page 323:
"Using pieces of genomes from first-born organisms to construct new genomes of later-born organisms." The different genomes for the first-born organisms were organized independently in the primordial pond resulting in unique organisms. When seed cells (and the individuals made from them) die, they break open, shedding their genomes into the primordial pond. Each of the broken DNA pieces from this genome, because it most likely contained some genes required for the construction of a living organism, had a far greater value than an equally-sized random DNA sequence in forming a genome for a multicellular organism. Therefore, such DNA pieces were bound to be used in the formation of new genomes along with other genes from the primordial pond. This would make some characteristics of the first-born organisms appear in later independently-born organisms.And this from figure 10.5 on page 489:
"Modification of an organism's genome in its free-living seed cell in the primordial pond and the production of a new creature." A seed cell capable of giving rise to a multicellular creature could reproduce for a long geological time and lose its special ability of growing into an organism while undergoing major modifications and repatterning in the genome. Rarely, a descendent seed cell in the primordial pond could give rise to a changed new organism, but with similarity to the organism produced from the starting seed cell.And finally, from the text on page 488:
This could lead to creatures changed slightly or considerably with similarities to the creature that first originated from the independently-assembled prototypical genome. ... Each independently-originating creature (whether invertebrate or vertebrate) also gave rise to many related similar species by many mechanisms such as natural selection and mutation, that is, by means of change through organismal descent with modification.Thus, the two trees you drew above for genes "X" and "Y are gross oversimplifications of the independent birth theory mechanism.
Arlin: (reprise) Will you also say that plants, animals and fungi are "too close" if I show congruence between them? This is a creationist ploy: all the best didactic examples of evolution are from closely-related things, where the signal-to-noise ratio is high and the evidence is clear.
Jeff: So, you chose closely-related vertebrates to improve the signal to noise ratio. However, as your own strict consensus tree shows, it is the most-distant organisms that show the least amount of noise. Tuna, Chicken, and mammals (as a group) have a better concensus (100%) than the creatures within the mammal group itself (less than 100%).
We're looking at a set of organisms and biomolecular sequence data. Our job is to decide if those organisms and data resulted from (1) descent from a common ancestor and/or (2) independent births from a pond. I submit that the organisms that we see today and your limited sequence data could come out the same using either path. To wit:
A: descent from a common ancestor:
organism #1 ----> mechanism "A" ----> organism #2
B: independent births:
organism #1 ----> mechanism "B" ----> organism #2
We need to determine, after the fact, which mechanism, A or B, is being used. Here are some of the characteristics of the mechanisms:
The two mechanisms look the same to me, except "A" cannot produce new and unique genes (according to Senapathy), and you assert that "B" cannot produce congruent trees, only common descent can -- but I have not agreed with that, and I suspect Senapathy does not either. I doubt we'll convince each other on the congruency argument.
Arlin: (reprise) This is not science. Science is being able to say what is consistent with the theory BEFORE SEEING THE DATA. Can you do this?
Jeff: I'm glad you said that, because it explains another reason why I didn't commit to your test. You knew what the data would show before you presented it to me, and it might be that this data is so often used in this way because it does support common descent, while contrary data is not pursued. I'm not accusing you of cooking the data, only that unfavorable data might not be coming to the forefront precisely because it does not support the only accepted theory around for the past 130 years. Also, the software program you used might have code in it that favors certain alignment attributes and not others -- I don't know -- but it would be reasonable to make the program behave that way because, after all, it is designed to show supposed alignments. I have the sources, but have not had time to examine them. We need to play by the same rules.
So, let's switch to another test, one which Senapathy has proposed himself (see test #1), and one were neither of us has prior knowledge of the data. That is, we should trace the origins of unique genes rather than similar genes. I asked you earlier about blood and immune system proteins, but you did not offer a very scientific answer to the origins of those genes.
You want me to make some sort of commitment beforehand. OK, you made the challenge, so I get to choose the weapons (one which I know Senapathy will agree to): let's look at unique genes found in supposedly "related" organisms. Can you explain how those unique genes came about, or propose a test (one which you do not have prior data for) to find out? If you'd rather use Senapathy's test #2 (DG pathways), that would be fine, too.
Although Senapathy does not mention transposons in his proposed tests, perhaps you could design a mutually agreeable test based on his statements on page 445:
"There are many transposons in the living world, and the transposons in various organisms are different. ... [F]or the evolutionary theory to be correct, the same transposons should be present in numerous creatures, or at least they should be the modified versions of a prototype, which would indicate an evolutionary relationship. ... It is highly improbably that only the tips of the evolutionary branches have developed unique transposons and sequences associated with them. ... If the ancestral organisms had some transposons then it is not possible for the descendent organisms to have abolished them and evolved new ones. ... [W]hen the gene sets for complex cells and organisms were randomly assembled, it is highly probably for a variety of distinct transposons to have assembled separately into different genomes."Maybe yet another test could be designed based on repetitive sequences, per the discussion on page 447.
[ar32]
Also, Arlin did a significant amount of "hand-waving" in trying to explain the relationship between invertebrate and vertebrate proteins, above. He didn't even attempt answer the question. Arlin said it best: "You are just saying things without trying to approach the truth" and it is "rubbish, magical, sky-hook explanation." I can find many such statements made by evolutionists.
Dr. Senapathy put it this way (at page 437):
The misinterpretations of evolutionists concerning similarity of genes are due to fundamental conceptual errors. These errors lead them to misinterpret the actual biomolecular scenario of similar functions, structures, and sequences of genes and proteins to be the result of organismal evolution. In addition, evolutionists also face several technical problems in searching for similar sequences in gene and protein sequence databases, misleading them to interpret false or chance similarity to be genuine similarity. The concept of evolution is strongly rooted in the minds of almost all scientists who begin their careers in the biological and biomedical sciences. The reason is that they are taught in school to believe that Darwin's theory is an established fact, and that only creationists oppose it for religious reasons. The minds of students are programmed to think that, scientifically speaking, evolutionary theory is correct. Because of this, they always look only for "evidence" for the evolutionary theory and ignore anything that is against the theory. Scientists practicing molecular biology are no exception. In their field, what they look for as evidence of evolution is similarity of proteins and genes in various organisms. To them, even new and unique genes evolve by evolutionary mechanisms of mutations. They assume that proteins and genes with different (i.e., unrelated) functions will have structural and sequence similarity. Therefore, when they find sequence similarity among proteins and genes that have unrelated functions, they group such proteins into a family of "heterofunctional proteins," implying that these evolved from a common ancestral gene from a common ancestral organism.
Dr. Senapathy discusses sequence similarities:
And in email to me, he writes:
Arlin's argument (above) is invalid, when he says that the alpha and beta globins were evolved from myoglobin, and the "gaps" that are similar in these molecules show that these organisms evolved from a common ancestor. It is true that these molecules have a lot in common, and it is possible that there is a relationship (or on the other hand, these molecules being so short, they could have occurred in independent random sequences also, as seen by the multiple occurrences of exons and genes in your interactive exon-search engine). But, even if these molecules are related, and were derived from one another, they could have done so in the primordial pond. You can see that the gaps that occur in one molecule occurs in all the organisms. It only means that all these organisms used both the genes (either from the primordial pool or from one original seed-cell), no matter by what mechanism these molecules came about (either by independent occurrences or by modification of one from another). The presence of the same gaps certainly does not mean any evolutionary relationship among these organisms.Also, I have not said that these molecules are totally distinct molecules, as Arlin says. I know very well that globins are the only and the most often quoted example for evolution today. They never talk about the distinct genes and proteins, because most of the biologists are even unaware of them.
Even if there is congruence among the differences that shows that these organism's genomes are related, it does not mean that these organisms are related by descent with modification. The basic reason for this argument is the presence of absolutely unrelated genes in distinct organisms such as the vertebrates and invertebrates, for which evolutionists do not have an answer. As you have correctly noted, the organisms that Arlin has used in his analysis are all vertebrates.
Back to [Part I]
Back to [Part II]